Virology and cellular imaging
Virology
Overview
Viruses are everywhere and hugely diverse, making them fascinating objects of study. As is clear from the SARS-Cov-2 pandemic, they also represent a threat to human health. For this reason, bio-medically relevant applications form an important motivation for our ongoing research in viral replication. These include the characterization of the methods of action of antiviral drugs as described above, the usage of force spectroscopy platforms for drug screening, and vaccinology.
Our recent contributions to virology have focused on:
- A better biophysical understanding of the mechanisms used by viruses to copy their genomes. This could allow us to develop new treatments against pathogenic viruses by impeding the correct and efficient replication of their genomes. Our research on viral replication focuses in particular on understanding the molecular processes that underline RNA replication of RNA-viruses, with the aim of gaining spatiotemporal insight into the molecular mechanisms of RNA-dependent-RNA polymerases (RdRp) responsible for carrying out RNA synthesis. In particular, we are interested in assessing how polymerases’ tendency towards misincorporation can be influenced by the presence of 1) RNA structures, 2) specific RNA sequences 3) and/or nucleotide analogs and, as such, provide ways of understanding viral adaptation and evolution, and ultimately contribute to the development of new antivirals.
- The biophysical properties of virus-like particles. Vaccines are currently the major means to prevent viral infection and global pandemics, yielding an imperative requirement for the development of safe, low cost, and scalable vaccine production process. A type of vaccine that has gained significant interest over the recent years, are virus-like particles (VLPs) vaccines, which are artificially synthesized, non-infectious simplified viral capsids that can provoke an immune response in a host. VLPs form a new, less costly approach in designing vaccines.
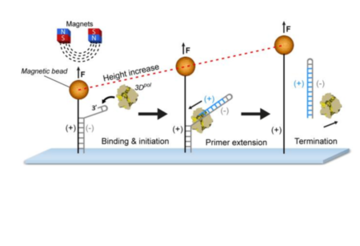
Monitoring synthesis by Viral Polymerase (RdRp)
Approaches
To examine nucleotide synthesis by viral RNA-dependent-RNA polymerases (RdRp), we use high-throughput single-molecule techniques, in particular magnetic tweezers. The high-throughput character of these techniques allows us to achieve high statistics and, in consequence, test quantitative models for polymerase mechanisms. Our assays consist of tracking tethered RNA molecules as they are converted from double-stranded RNA to single-stranded RNA by a single RNA-dependent RNA polymerase. Due to the applied force on the bead, and the fact that single-stranded DNA is more extensible than the double-stranded helix, we see this as a length change over time. This can then be converted into a number of RNA nucleotides synthesized over time. Studying RdRp elongation dynamics permits the direct observation of the behavior of the polimerases that can include copy-back synthesis, back tracking, stability of the RdRp-nascent-RNA complex, and the dimensions of the RdRp nucleic-acid-binding channel.
To examine the biophysical properties of VLPs, we have used a diverse set of biophysics tools such as AFM, and (cryo)-EM, as well as various biochemical approaches to characterise their contents.
Recent findings
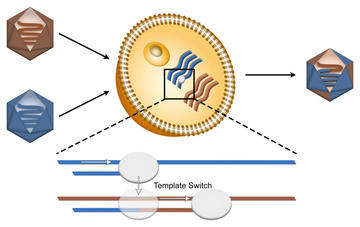
Illustration of viral recombination. Our work on EV-A71 RdRp illustrates that a combination of backtracking and polymerase flexibility leads to an enhanced incidence of both copy-back synthesis and recombination in EV-A71 virus.
We have used magnetic tweezers to study the RdRp of EV-A71 virus, and shown that this RdRp is particularly prone to copy-back synthesis and rrecombination, which derive from similar mechanisms. The figure at right shows the recombination pathway. The occurrence of both phenomena was also increased when we added an antiviral, T-1106-TP. This observation highlights a previously unknown mechanistic impact of antivirals, and may provide further means of impacting viral proliferation.
Our recent work on the biophysical properties of EV-A71 VLPs will be described here shortly.
Researchers involved
- Richard Janissen
- Louis Kuijpers
- Belén Solano
Collaborators
- Craig Cameron (Penn State University, USA)
- Shin-Ru Shih (Chang Gung University, Taiwan)
- Marco Vignuzzi (Institut Pasteur, France)
- Arjen Jakobi (TU Delft)
- Wouter Roos (Groningen University)
- Leo van der Pol (Intravacc, Dutch Institute of Vaccinology)
Cellular Imaging
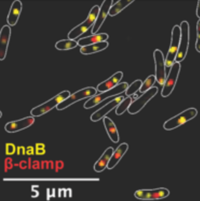
While most of our research in conducted in vitro, using reconstituted systems, of course the true context of biological processes is the living cell. We have previously examined various aspects of bacterial replication in the cellular context. Most recent student projects have focused on studying the spatial-temporal impact that DNA-damaging drugs and UV radiation have on the different components of the replisome, and on accessory helicases, in live cells of bacterium E. coli These projects made use of genetic engineering; live cell, single-molecule fluorescence, confocal and super-resolution microscopy approaches; and where necessary, microfluidic devices. We will be exploring similar approaches together with various other Oxford groups.



