Eukaryotic chromatin replication
Research overview
DNA replication is carried out at very high accuracy by nanometer-scale, multi-protein complexes known as replisomes. In eukaryotic organisms such as ourselves, the replisome consists of some twenty different proteins. Eukaryotic replication occurs in the context of chromatin: the meters of DNA in eukaryotic organisms are tightly packed into a higher-order structure called chromatin in order to fit in the tiny nucleus. The basic compaction unit of this condensed structure is a DNA-protein complex termed nucleosome which consists of a small piece of DNA wrapped around a core of so-called histone proteins. This compaction adds an extra layer of complexity to the replication process.
Our research focuses on understanding the molecular processes that underlie eukaryotic DNA replication in the context of chromatin, with the particular aim of gaining spatiotemporal insight into their dynamics by using our single-molecule biophysical expertise in replication and chromatin while integrating it with state-of-the-art molecular biology and biochemistry.
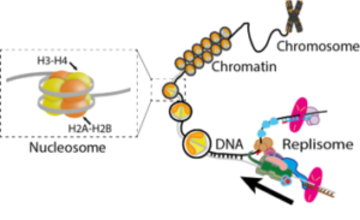
Schematic of DNA replication in the context of chromatin. The eukaryotic replisome copies DNA that is wrapped about nucleosomes. This state of DNA is also known as chromatin.
Approach
Despite tremendous advances in understanding chromatin replication achieved by experiments in genetics, cell biology, structural biology, and biochemistry, a detailed mechanistic understanding of how the replisome interacts with nucleosomes, histone chaperones, and chromatin remodelers still remains. The possibility of reconstituting an active yeast replisome in vitro in our lab (originally described by the Diffley laboratory in 2015) has opened up a new perspective, because when integrated with reconstituted forms of chromatin it provides the means to study chromatin replication in a precise, carefully controlled manner.However, to really understand how the different processes that maintain robust chromatin replication occur in space and time requires probing the stoichiometry and dynamics of the individual proteins involved. Doing so requires a complementary approach to bulk biochemistry that can be found in single-molecule techniques. These high-resolution techniques, which include single-molecule fluorescence and single-molecule force spectroscopy, monitor individual biochemical processes under physiological conditions in real time and have demonstrated their value in revealing the dynamics of large protein complexes.
Recent work
In our latest study, we’ve ventured into the dynamic world of ORC proteins and their behavior in the presence of nucleosomes, the structural units of chromatin. Here are the key findings: 1. Dynamic ORC protein: ORC in yeast is a highly dynamic protein. It moves rapidly along DNA, but its journey can be halted when it encounters specific sequences known as “origin recognition sequences.” 2. Chromatinized Origins: We’ve explored the dynamics of ORC proteins in the context of chromatinized origins, where nucleosomes are present. Surprisingly, we’ve found that ORC efficiently binds to the origin of replication, whether it’s chromatinized or not. However, the presence of nucleosomes reduces the mobility of ORC. 3. Efficient MCM Recruitment: The recruitment of MCMs to chromatinized origins is very efficient. 4. MCM Retention: On chromatinized origins, MCM faces severe constraints in moving away from the origin. This suggests that chromatinized origins play a vital role in retaining MCM locally, potentially setting the stage for the assembly of the replisome, the molecular machinery responsible for DNA replication. These findings shed light on the intricate relationship between chromatin structure, ORC dynamics and MCM loading, deepening our understanding of the mechanisms underlying DNA replication. Stay tuned for further developments as we continue to explore the fascinating world of chromatin and DNA replication!
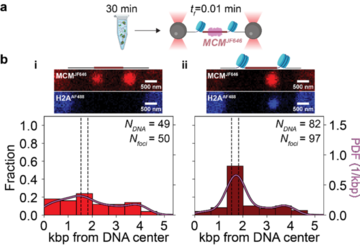
Nucleosomes spatially constrain MCM at the origin. (a) ORC, Cdc6, and MCMJF646/Cdt-1 interact with 10.4 kb DNA molecules (chromatinized or not) for 30 min with ATP. The DNA-protein complex is then transferred to a single-molecule flow cell, tethered, and scanned at an acquisition rate of one frame every 0.01 min (tf = 0.01 min). (b) Schematics of origin regions, representative full-length fluorescence scans (red: MCMJF646; blue: H2AAF488), and spatial distributions of MCMJF646 immediately after introduction into the flow cell. (i) MCMJF646 distribution on DNA with a non-chromatinized origin based on diffraction-limited red spots (Nfoci) from 44 DNA molecules (NDNA-MCM). Dashed lines show nucleosome positioning locations, and the solid curve is a kernel density estimation (PDF: probability density function). (ii) MCMJF646 distribution on DNA with a chromatinized origin based on diffraction-limited red spots (Nfoci) from 77 DNA molecules (NDNA-MCM).
Openings for students
We typically have several projects available, please contact us to find out the latest.
Researchers currently involved in these projects
- Zhaowei Liu
- Yasemin Baris
- Francisco Palmero Moya
- Hoa Vo
- Kamila Myka
- Belén Solano
- Together with our former Delft colleagues Pang Yen Wang and Theo van Laar



