Eukaryotic DNA replication
Research overview
The copying, or replication, of DNA is one of the central processes that take place in all living organisms. Our understanding of DNA replication has made gigantic leaps forward since the discovery of the double helical form of DNA by Watson and Crick in 1953. We know many of the structures and functions of the proteins and enzymes involved. Many of these discoveries have been made by studying DNA replication in simple systems, such as viruses or bacteria. These continue to yield valuable insights, but recently, advances in the reconstitution of the yeast replisome have made it possible to gain insights into eukaryotic replication. DNA replication is carried out at very high accuracy by nanometer-scale, multi-protein complexes known as replisomes. In eukaryotic organisms such as ourselves, the replisome consists of some twenty different proteins.
Our research focuses on understanding the molecular processes that underlie eukaryotic DNA replication, with the particular aim of gaining spatiotemporal insight into their dynamics by using our single-molecule biophysical expertise in replication while integrating it with state-of-the-art molecular biology and biochemistry.
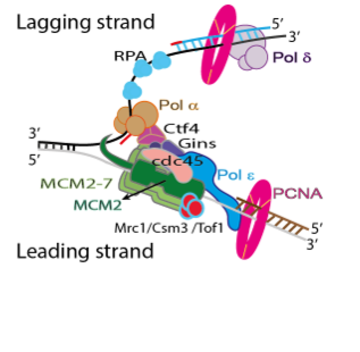
Schematic of the eukaryotic replisome. The central motor of the replisome is the CMG helicase, which unwinds the parental DNA; the polymerases are then able to add new nucleotides on the leading and lagging strands, respectively, leading to two daughter DNA molecules.
Recent works
Dynamics of replisome loading proteins
We have recently published our findings about the dynamics of the loading proteins involved in the first step of replisome assembly. The origin recognition complex (ORC) turns out to be a protein that is quite mobile on the DNA, except at the origin. Recruitment of the MCM helicase in an ORC-dependent manner can occur at different locations on the DNA, but immobile ORC-MCM complexes are also preferentially observed at the origin. When loaded onto DNA in the presence of ATP in bulk experiments and then visualized at the single-molecule level, both single and double Mcm2-7 hexamers are found on the DNA, and both exhibit similar low-level mobility.
Dynamics of replicative CMG helicase
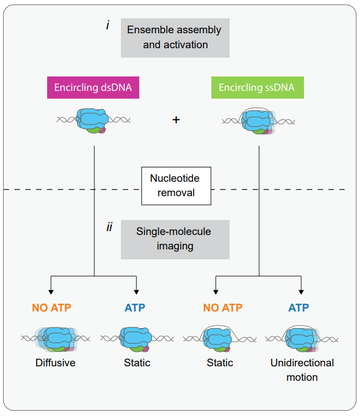
We reconstituted the assembly and activation of the replicative helicase CMG from 36 polypeptides, and for the first time imaged and quantified its motion at the single-molecule level using a combination of dual-bean optical tweezers and confocal microscopy. To this end, we developed a novel hybrid ensemble and single-molecule assay relying on the dual
functionalization of the DNA substrate with two orthogonal attachment types. This assay allowed us to assemble and activate CMG in bulk, purify the resulting CMG:DNA complexes from excess unbound protein, and image the purified product at the single-molecule level.
Our single-molecule imaging showed that CMG moves unidirectionally in the presence of ATP, consistent with previous work from other groups. Surprisingly, however, we observed that CMG can also move in the absence of ATP, but that it does so in a diffusive manner. Furthermore, we show that ATP binding halts this diffusive motion.
To explain our findings, we propose a model that’s summarized in the figure on the right. We propose that the ATP binding-mediated halting that we observe keeps CMG close to replication origins and facilitates the initial DNA melting required to initiate DNA replication.
Openings for students
We typically have several projects available, please contact us to find out the latest.
Researchers currently involved in these projects
- Saurabh Talele
- Yasemin Baris
- Francisco Palmero Moya
- Kamila Myka
- Zhaowei Liu
- Belén Solano
- together with our former Delft colleagues Theo van Laar and Daniel Ramírez Montero
Current collaborators
- John Diffley Lab (Francis Crick Institute, UK)
- Alessandro Costa Lab (Francis Crick Institute, UK)
- Thomas Miller Lab (University of Copenhagen, DK)
- Hasan Yardimci Lab (Francis Crick Institute, UK)
- We are also grateful to e.g. the Joe Yeeles lab at the LMB for providing us with strains.



