Research overview
Our research focuses on understanding the molecular processes that underlie DNA and RNA replication, with the particular aim of gaining spatiotemporal insight into their dynamics by using our single-molecule biophysical expertise in replication and chromatin and integrating it with state-of-the-art molecular biology and biochemistry.
While the primary aim of our research is to give fundamental mechanistic insight into different aspects of replication, several bio-medically relevant applications also form part of our ongoing research. These include the usage of force spectroscopy platforms for drug screening and vaccinology.
Why is this a challenge?
The copying, or replication, of DNA is one of the central processes that take place in all living organisms. DNA replication, essential to accurately transmit all genetic information to daughter cells upon cell division, is a key process to cellular function, proliferation, and survival. When the replication process is compromised, DNA damage and mutations will start to accumulate. For some organisms such as viruses or bacteria, error-prone replication can be their benefit, for example by creating more virulent viral strains, allowing viruses to jump from animal to human species (zoonosis), or allowing bacteria to develop antibiotic resistance. However, for humans and other species, deficient DNA replication will be a major source of the mutations and chromosome rearrangements that accumulate over a lifetime, resulting in pathological disorders such as cancer and aging.
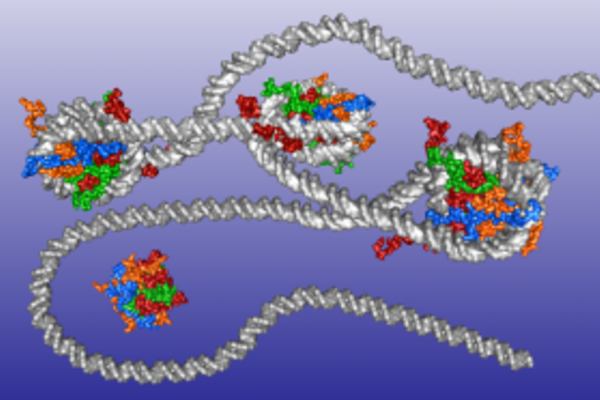
The process of replication is orchestrated by the assembly and activation of the replisome, a complex macromolecular machine that contain multiple components that transiently associate with each other during replication. The replisome is generally most complex in higher eukaryotes, and simplest in viruses, where a polymerase may sometimes suffice. Still, there are a number of viruses where the activity of multiple proteins is necessary to carry out replication, including e.g. SARS-CoV-2. In eukaryotes, while advances in biochemistry and structural biology have led to the identification of the essential roles played by the different replisome components, and an overall timeline for their interaction has been constructed, describing the complex dynamic interplay between them, particularly at short time scales, still presents a challenge.
Research topics
Our research projects are grouped in three main topics: eukaryotic DNA replication , eukaryotic chromatin replication, and viral and bacterial replication . Some current projects are listed below:
Some of our current projects include:
- Monitoring the assembly of the yeast eukaryotic replisome on chromatin and visualize how nucleosomes and other roadblocks impact its progression.
- Monitoring the interplay between yeast eukaryotic replisome components and chaperones to study the coupling of replication to nucleosome assembly and reassembly during replication
- In live bacterial cells, studying the spatial-temporal impact that DNA-damaging drugs and UV radiation have on the different components of the replisome and on accessory helicases.
- Describing the method of action of different antiviral agents by applying our force spectroscopy methods to viral replication.
Approach
To reach this level of understanding, it is required the observation of the workings of individual components of the replisome in real time during replication, and this can only be achieved by using single-molecule techniques. Indeed, these techniques allow us to look at replication in a specific time-window, thereby capturing short-lived states that are normally not visible in ensemble assays but which can contain essential information to fully decipher the workings of the replisome.